Tutorial: using NeurEco Python API on a Tabular Compression problem
Tutorial: using NeurEco Python API on a Tabular Compression problem#
The following section uses the test case Heaviside. This test case is included in the NeurEco installation package.
Create an empty directory (Heaviside Example), extract the Heaviside test case data there. The created directory contains the following files:
x_test.csv
x_train.csv
Import the required libraries (NeurEco and NumPy):
from NeurEco import NeurEcoTabular as Tabular
import numpy as np
Load the training data:
x_train = np.genfromtxt("x_train.csv", delimiter=";", skip_header=True)
Initialize a NeurEco object to handle the Compression problem:
builder = Tabular.Compressor()
All the methods provided by the Compressor class, can be viewed by calling the __method__ attributes:
print(builder.__methods__)
**** NeurEco Tabular Compressor methods: ****
- load
- save
- delete
- evaluate
- build
- get_input_count
- get_output_count
- load_model_from_checkpoint
- get_number_of_networks_from_checkpoint
- get_weights
- export_fmu
- export_c
- export_onnx
- export_vba
- compute_error
- separate_models
- concatenate_models
- plot_network
- plot_compression_coefficients
- forward_derivative
- gradient
- set_weightsplot_compression_coefficients
- perform_input_sweep
To understand what each parameter of any method does and how to use it, print the doc of the method:
print(builder.export_c.__doc__)
exports a NeurEco tabular model to a header file
:param h_file_path: path where the .h file will be saved
:param precision: string: optional: "float" or "double": precision of the weights in the h file
:return: export_status: int: 0 if export is ok, other if otherwise.
To build the model, run the build method with the building parameters adjusted to the problem at hand (see Build NeurEco Compression model with the Python API). For this example, the outputs are normalized per feature (meaning that each output is normalized apart, it is the default setting for Compression, see Data normalization for Tabular Compression):
builder.build(x_train, # the rest of the parameters are optional
write_model_to='./HeavisideModel/Heaviside.ednn',
write_compression_model_to='./HeavisideModel/HeavisideCompressor.ednn',
write_decompression_model_to='./HeavisideModel/HeavisideUncompressor.ednn',
compress_tolerance=0.050,
checkpoint_address='./HeavisideModel/Heaviside.checkpoint',
final_learning=True,
initial_beta_reg=0.1)
When build is called, NeurEco starts the building process:
Validation Percentage will be used to get the validation data. This is due to:
- one or all the validation data is set to None
- validation indices is set to None
info >
info > _ __ ______
info > / | / /__ __ _______/ ____/________
info > / |/ / _ \/ / / / ___/ __/ / ___/ __ \
info > / /| / __/ /_/ / / / /___/ /__/ /_/ /
info > /_/ |_/\___/\__,_/_/ /_____/\___/\____/
info > === A D A G O S ===
info >
info > Version: 4.01.2474.0 Compiled with MSVC v1928 Oct 12 2022 Matlab runtime:no
info > OpenMP: yes
info > MKL: yes
info > Reading data files...
info > Reading Data from C:/Users/Sadok/AppData/Local/Temp/tmpy1bh9n82/inputs_tab_comp_train.npy
info > build for: 20 outputs and 20 inputs and 400 samples.
During the build NeurEco saves the intermediate modes to the checkpoint file (defined by the parameter checkpoint_address). To load and use the intermediate models from this checkpoint:
Create a new NeurEco object in which to load the model:
model = Tabular.Compressor()
Determine how many intermediate models the checkpoint contains:
n = model.get_number_of_networks_from_checkpoint("./HeavisideModel/Heaviside.checkpoint")
Load any intermediate model from the checkpoint using its id (count starts with zero). For this example, at the moment of running the command \(n=2\) and the following command loads the intermediate model \(n°1 \ (id=0)\):
model.load_model_from_checkpoint("./HeavisideModel/Heaviside.checkpoint", 0)
Now model is a valid Compression model, and can be used as usual.
Check the number of trainable parameters each of the intermediate models has:
for i in range(n):
print("Loading model", i, " from checkpoint file:")
model.load_model_from_checkpoint("./HeavisideModel/Heaviside.checkpoint", i)
print("number of trainable parameters in intermediate model --", i, " is:", model.get_weights().size)
Loading model 0 from checkpoint file:
number of trainable parameters in intermediate model -- 0 is: 676
Loading model 1 from checkpoint file:
number of trainable parameters in intermediate model -- 1 is: 1220
Loading model 2 from checkpoint file:
number of trainable parameters in intermediate model -- 2 is: 1731
Loading model 3 from checkpoint file:
number of trainable parameters in intermediate model -- 3 is: 2145
Loading model 4 from checkpoint file:
number of trainable parameters in intermediate model -- 4 is: 2157
Loading model 5 from checkpoint file:
number of trainable parameters in intermediate model -- 5 is: 2157
Once the build is over, we can move to evaluating it on new data. To do so, we will start by separating it into a compression model and a decompression model, which are both regression models in this case. This is done by either loading them from the disk separately (in the build we asked for each model to be saved separately), or we can call the method separate_models of a NeurEco Compressor. We will load the model first:
Create a Compressor object to use for the evaluation:
combined_model = Tabular.Compressor()
Note
It is possible to use the already existing Compressor object builder when the evaluation is done just after the build, and builder is still available.
Load the built model:
combined_model.load("./HeavisideModel/Heaviside.ednn")
To separate the Compressor model into two parts: a Regressor model for compression part and a Regressor model for the decompression part:
Create two new Regressor objects to use:
neurEco_Compressor = Tabular.Regressor() neurEco_Decompressor = Tabular.Regressor()
Separate the Compressor model into a compressor and a decompressor:
separate_status = combined_model.separate_models(neurEco_Compressor, neurEco_Decompressor)
Note
When building or evaluating a NeurEco model, all the used paths do not necessarily need to have an extension when they are passed as parameters to a NeurEco method.
Evaluate the separated models on the testing data and compute the compression error:
compressed_coefficients = neurEco_Compressor.evaluate(x_test)
decompressed_output = neurEco_Decompressor.evaluate(compressed_coefficients)
compression_error = neurEco_Decompressor.compute_error(decompressed_output, x_test)
print("The non-linear compression error is (%):", 100 * compression_error)
The non-linear compression error is (%): 1.898570291522237
Note
The relative compression error is highly dependent on the tolerance chosen for the build. A smaller tolerance leads to a more complex model and a better error on the testing set and the training set.
During evaluation, the normalization is carried out by the model and its parameters are not relative to the data set being evaluated, but are the global parameters computed during the build of the model.
The obtained result decompressed_output is the same as the output of the call:
decompressed_output_combined = combined_model.evaluate(x_test)
The neurEco_Compressor and neurEco_Decompressor models can be used as regular Regression models. For example, to extract the information about neurEco_Compressor, run:
n_inputs = neurEco_Compressor.get_input_count()
n_outputs = neurEco_Compressor.get_output_count()
weights = neurEco_Compressor.get_weights()
print("Number of inputs for the compression model:", n_inputs)
print("Number of nonlinear coefficients:", n_outputs)
print("Number of trainable parameters - compression block:", weights.size)
Number of inputs for the compression model: 20
Number of nonlinear coefficients: 2
Number of trainable parameters - compression block: 901
Plot the network graph (see Plot a NeurEco network, this operation requires matplotlib library installed) for any model (compressor, decompressor or combined):
combined_model.plot_network()
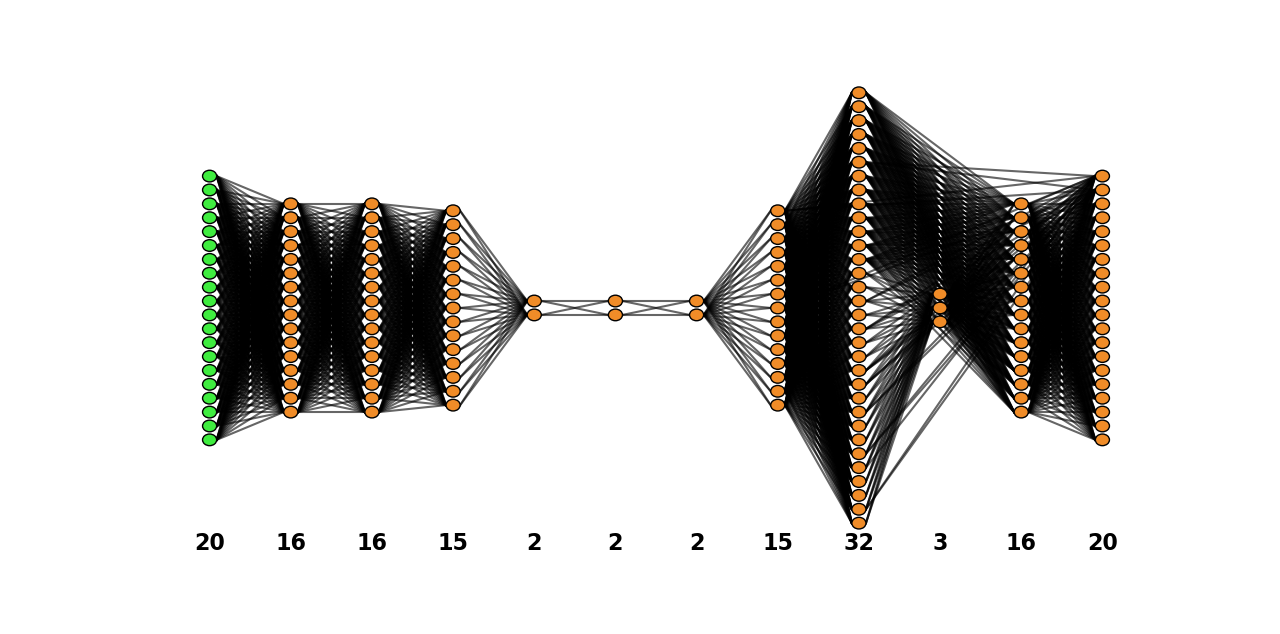
Python API operations: plotting a network: test case - Heaviside#
Plot the compression coefficients (this operation requires matplotlib library installed).
For this test case (Heaviside), one of the parameters (\(a1\)) represents the amplitude of the signal. All the amplitudes are contained in the interval [-0.6, 1]. Let’s get all the amplitude higher than 0.2 and all the amplitude lower than 0.2 separated (0.2 is the middle point of the interval).
amplitudes = []
for i in range(x_train.shape[0]):
sample = x_train[i, :]
diff = np.diff(sample)
amplitudes.append(np.max(sample))
amplitudes = np.array(amplitudes)
high_amplitudes_indexes = np.where(amplitudes > 0.2)[0]
low_amplitude_indexes = np.where(amplitudes < 0.2)[0]
Create a binary set of classes “h” and “l” for higher and lower, and plot the compression coefficients for these classes.
classes = np.empty(x_train.shape[0]).astype(str)
classes[high_amplitudes_indexes] = "h"
classes[low_amplitude_indexes] = "l"
combined_model.plot_compression_coefficients(data_to_compress=x_train, neurons_ids=[0, 1], data_labels=list(classes))
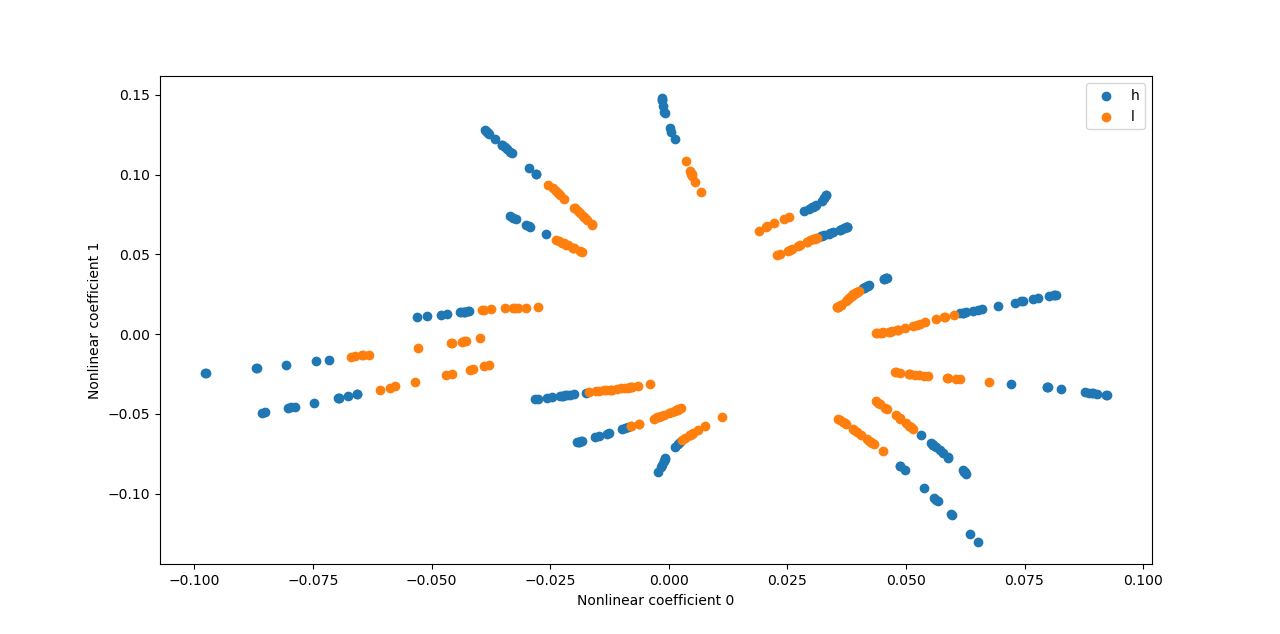
Python API operations: Plotting the nonlinear coefficients: test case - Heaviside#
To perform an input sweep (see Input sweep, this operation requires matplotlib library installed), run, for example:
combined_model.perform_input_sweep(x=x_test[43, :], input_id=2, input_interval=[-1, 1], output_id=0)
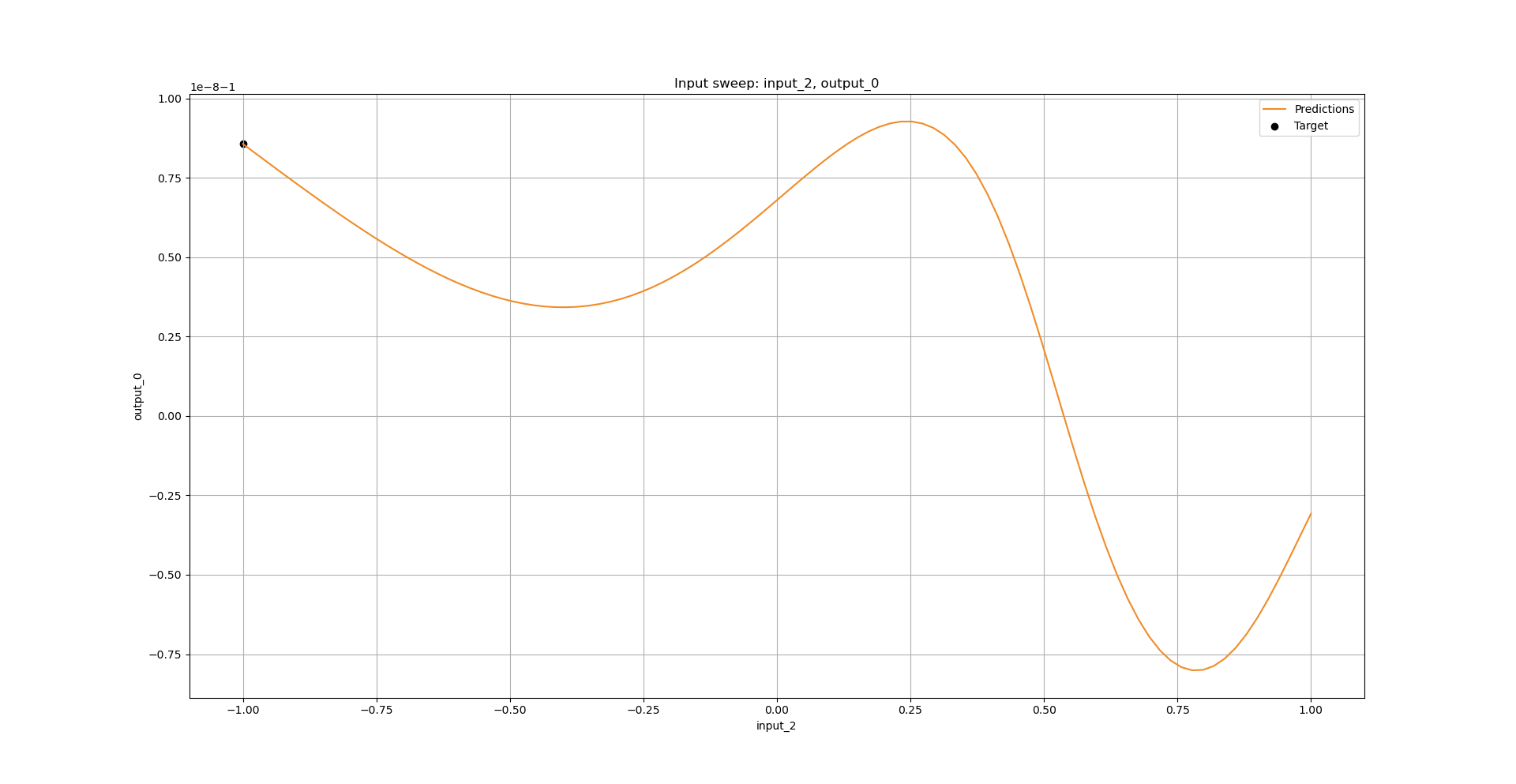
Python API operations: Performing an input sweep: test case - Heaviside#
To save the model in the native NeurEco binary format:
save_state = combined_model.save("Heaviside/NewDir/SameModel")
To export the model, run one of the following commands (embed license is required):
combined_model.export_c("./HeavisideModel/Heaviside.h", precision="float")
combined_model.export_onnx("./HeavisideModel/Heaviside.onnx", precision="float")
combined_model.export_fmu("./HeavisideModel/Heaviside.fmu")
combined_model.export_vba("./HeavisideModel/Heaviside.bas")
Note
The compressor and decompressor parts of the combined_model are exported automatically as well.
Warning
Once the NeurEco object is no longer needed, free the memory by deleting the object by calling the delete method. For the example above, five objects must be deleted
builder.delete()
combined_model.delete()
model.delete()
neurEco_Compressor.delete()
neurEco_Decompressor.delete()
